User's manual
Table of contents
Introduction
Index page
Browse page
Family entry page
Text search page
Protein search page
Domain search page
Detailed view of an entry
Database download page
Introduction
Transmembrane proteins constitute approximately 20-30% of fully sequenced proteomes and are responsible for a wide variety of cellular functions.
They can be divide into two main structural categories, the alpha-helical membrane proteins and the beta-barrels.
The former are located primarily in cell membranes of eukaryotic cells and bacterial inner membranes, while the latter are found exclusively in the outer membranes of Gram-negative bacteria, and presumably in the outer membranes of mitochondria and chloroplasts.
Currently, there is only a small number of available high resolution 3D-structures of ß-barrel outer mebrane proteins (OMPs) and this intensifies the need for accurate and comprehensive data collection regarding their topology.
OMPdb offers the largest dataset of OMPs worldwide and aims at bridging the gap between incomplete and/or false annotation of such proteins that is found in other public databases.
Return to top
Database organization
Index Page
In the welcome (index) page of OMPdb's website, the user can see a general description of the database, the current holdings and the date of its last update. From the drop-down menu on the right of the page, named Detailed view of an OMPdb family entry, the user can view the desirable family entry. Further, the database can be searched with a Uniprot accession number (Enter a Uniprot accession number textbox) directly.

Return to top
Browse
OMPdb is structured around the "Family" Concept. When the user clicks on the Browse page, he/she is presented with the available functional classes in which the OMPdb families and their members belong to. By clicking on each of the classes tabs, a drop-down menu appears, from which the user can navigate to each of the families, for further information.
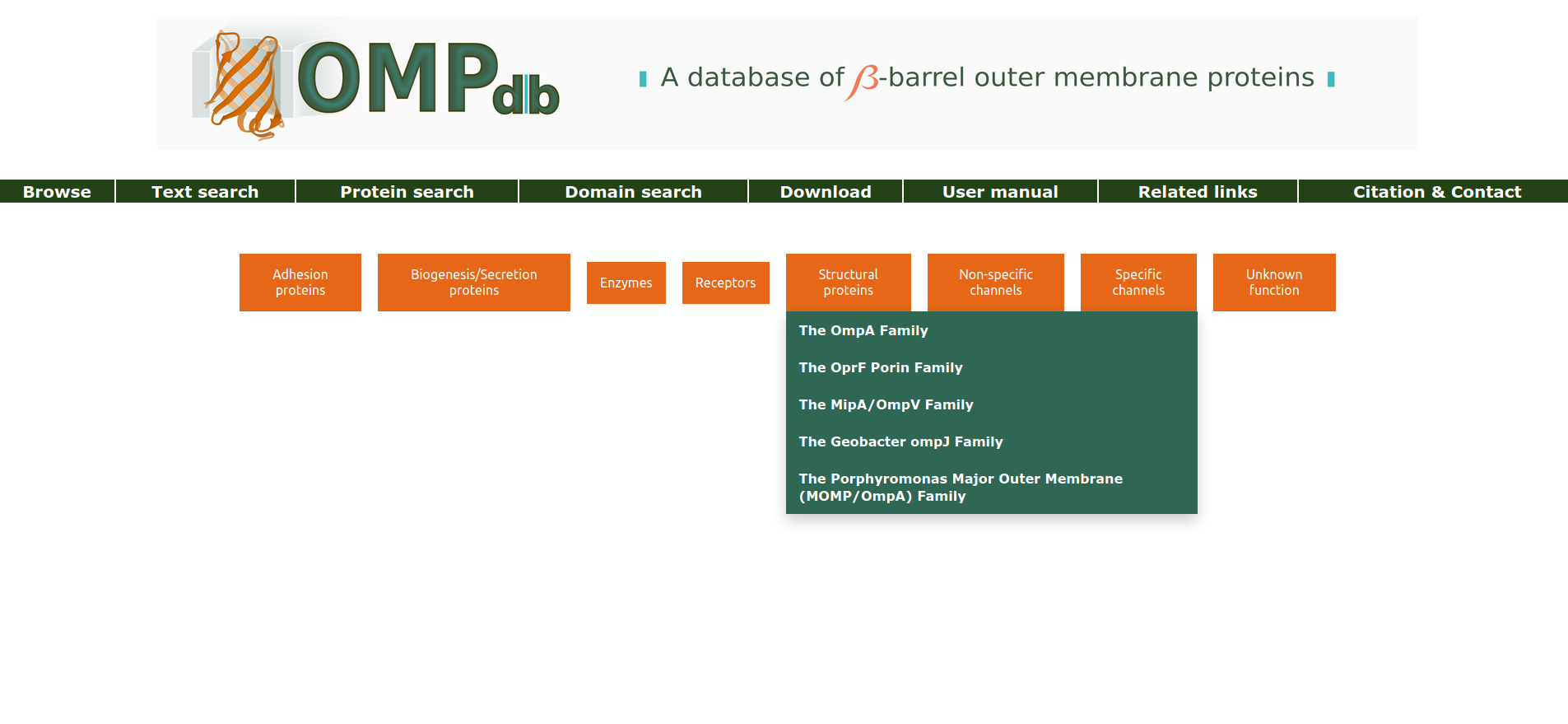
Return to top
Family entry
In the family entry page, the user can view its description, along with literature references related to the creation of the family.
There is also the oppurtunity to view all proteins in OMPdb that belong to this family. The user can retrieve all proteins that belong to the respective
family, or just those that match more than a specified coverage of the profile HMM model that characterizes the family. Finally, a list of proteins
that have an experimentally determined 3D-structure, is included, if applicable.
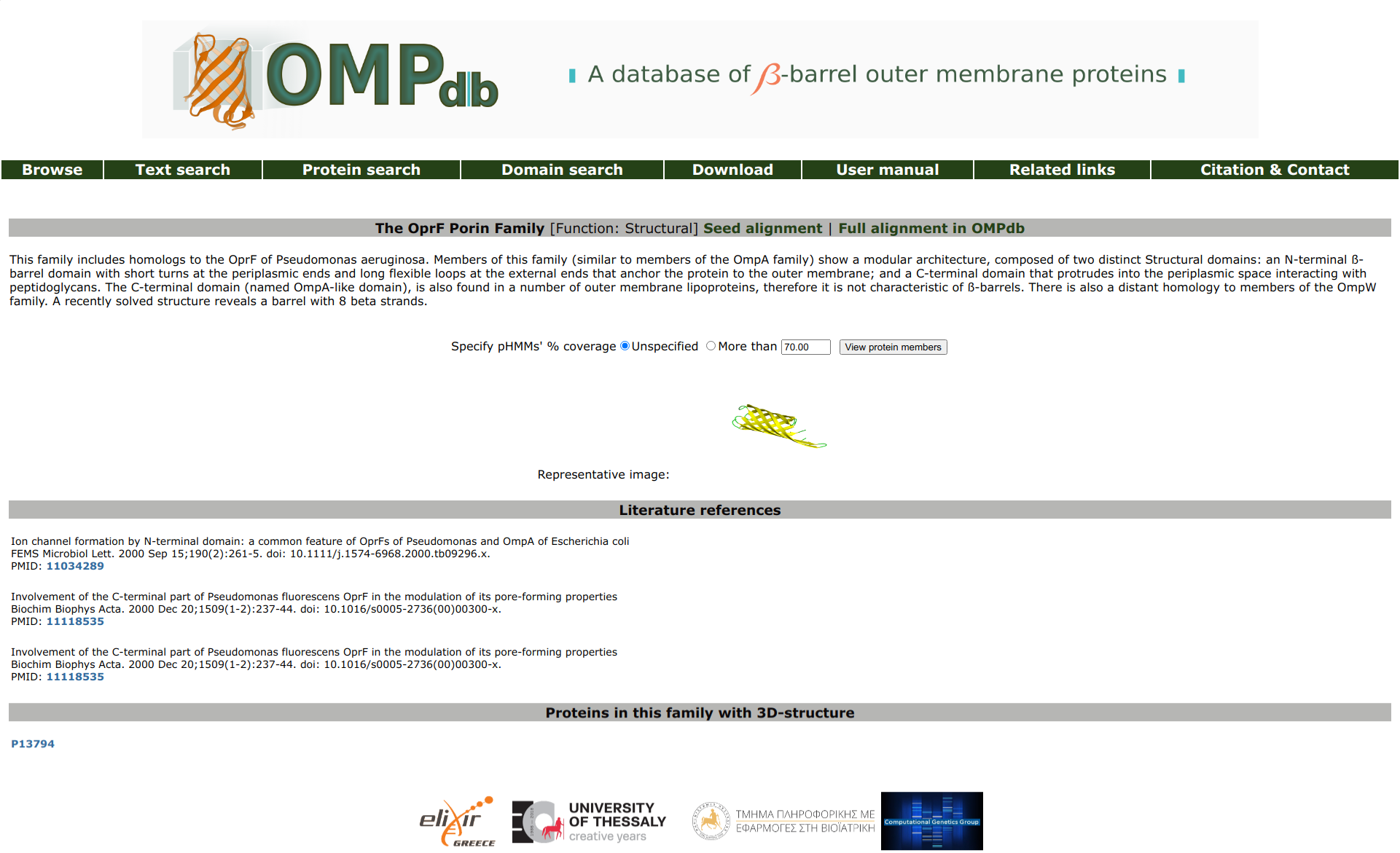
Return to top
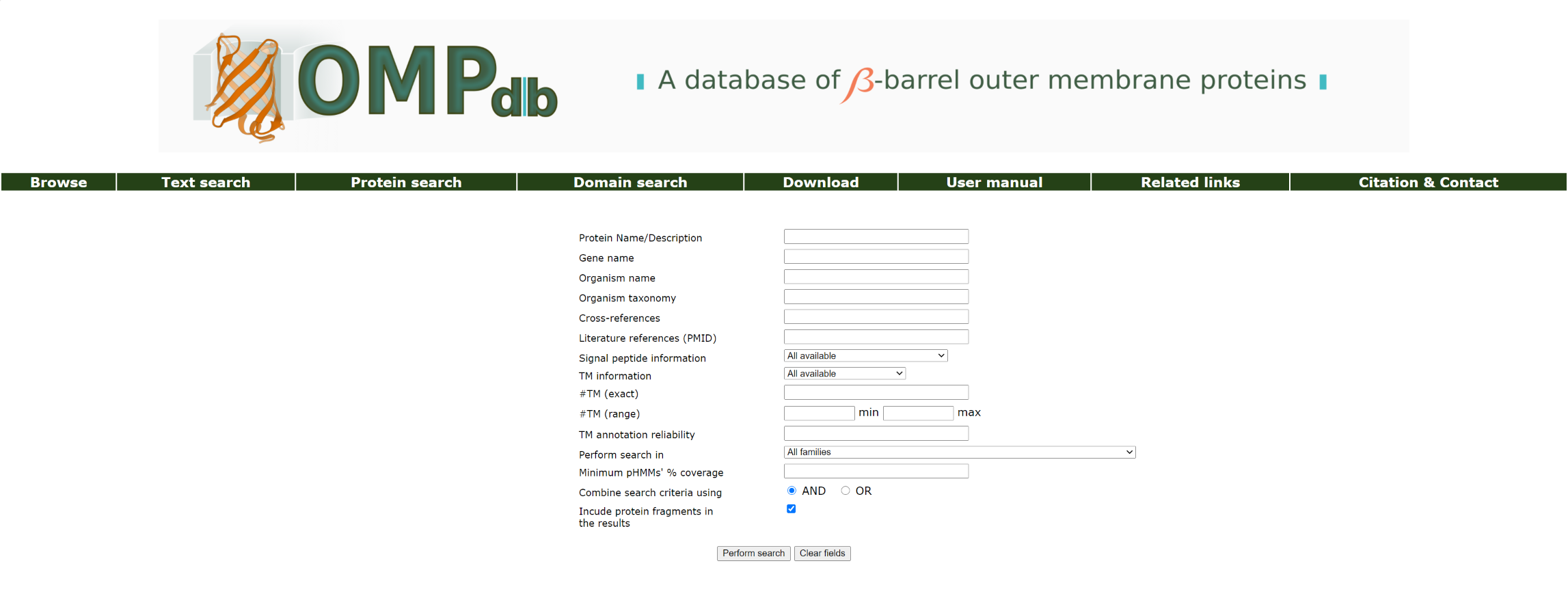
Text Search
In the Text Search page, the user can search for any text in the fields of his/her preference. The user can enter any word (or expression) in one or more of the available textboxes, namely Protein Name/Description, Gene name, Organism name, Organism taxonomy, Cross-references and Literature references (PMID). In the Signal Peptide information field, the user can choose to include one of the available types of information concerning the existence of a signal peptide in the search, while from the drop-down menu below (Perform search in), a family within the search will be limited to can be selected. The exact number (#TM (exact))or a range (min-max - #TM (range)) number of beta strands can be entered as well, together with a reliability about this annotation (TM annotation reliability). A drop-down menu with the TM annotation source can be found under TM information. The desired % coverage of the profile HMM models can also be specified using the radio-button Specify pHMMs' coverage (optional).
The Combine search criteria using field allows the user to decide if all or at least one of the several sub-expressions he/she may have entered in the text search fields will be combined for the results retrieval (using AND operator or OR operator respectively). The user has also the ability to select if protein fragments (based on the Uniprot description of the protein entry) will be included in the results or not (Incude protein fragments in the results checkbox).
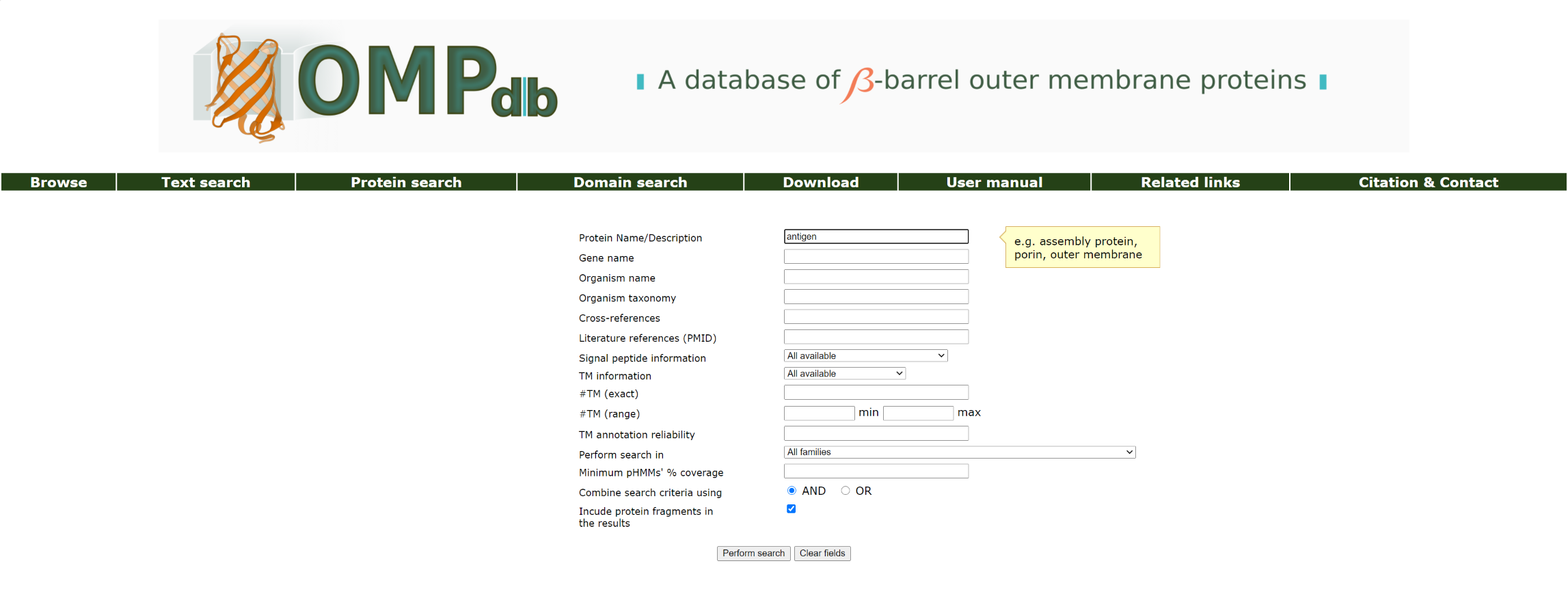
Protein name
Corresponds to the field Protein description of an entry.
Example 1:
If the user wants to retrieve all proteins that have the word antigen in their description, he/she must enter the keyword antigen as a query in the Protein name field as shown below:
At the result's page, the user can see a short description of the entries matching the search criteria and visit each of the available entries he/she wishes, by clicking on one of the Uniprot ACs links under View entry.
The top of the result page for the above search is:
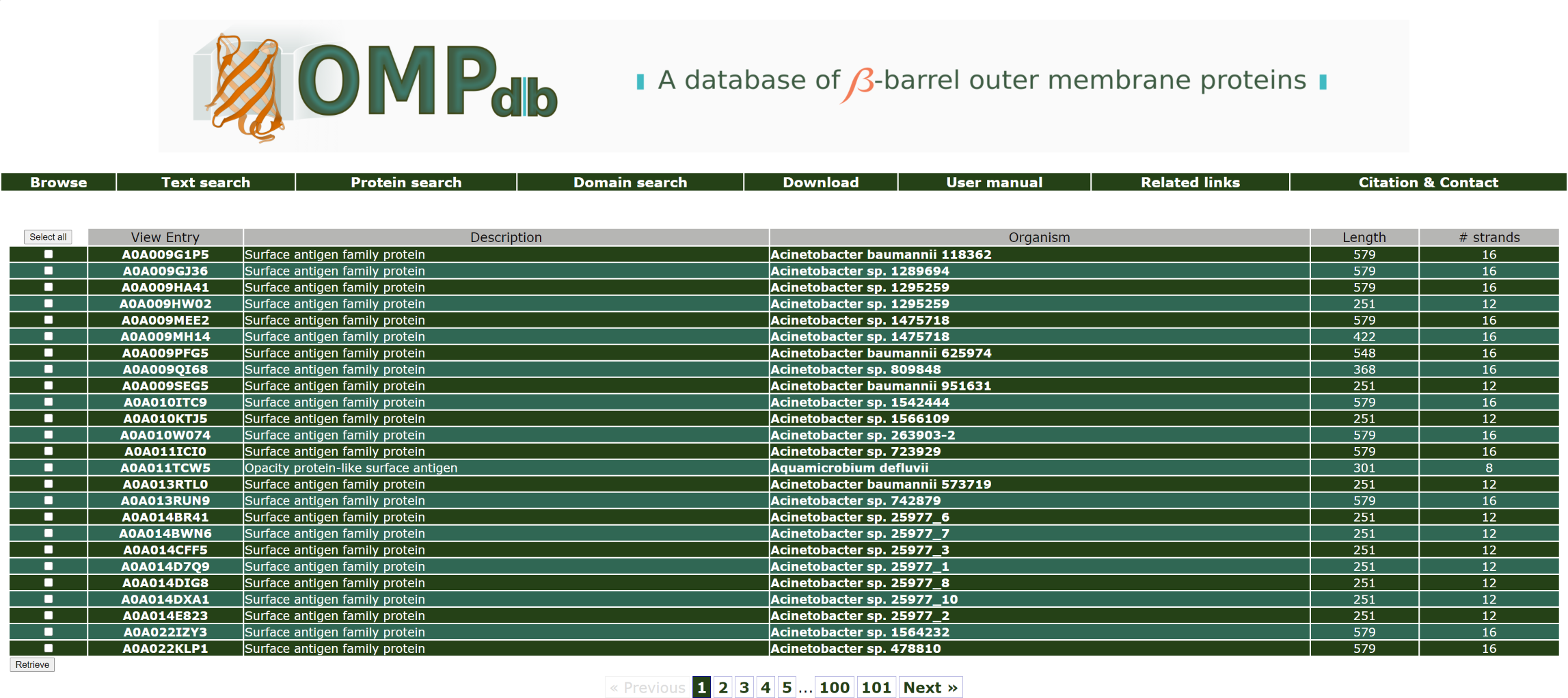
! Notice that the user can click on an Organism name in order to retrieve all proteins in the database from this specific organism.
Return to top
Gene name
Corresponds to the field Gene name of an entry.
Example 2:
If the user wants to retrieve all proteins with genename similar to ompL1 that are included in OMPdb, he/she should enter ompL1 as a query in the Gene name field:
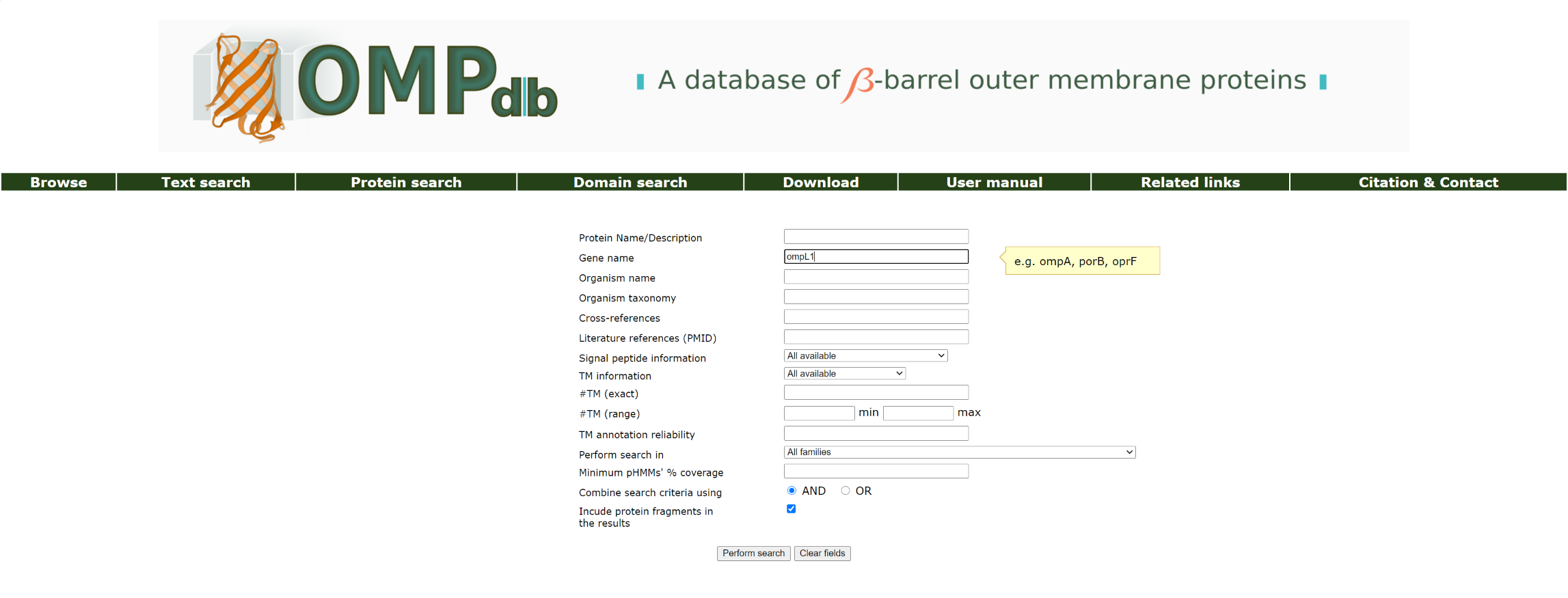
The top of the result page for the above search is:
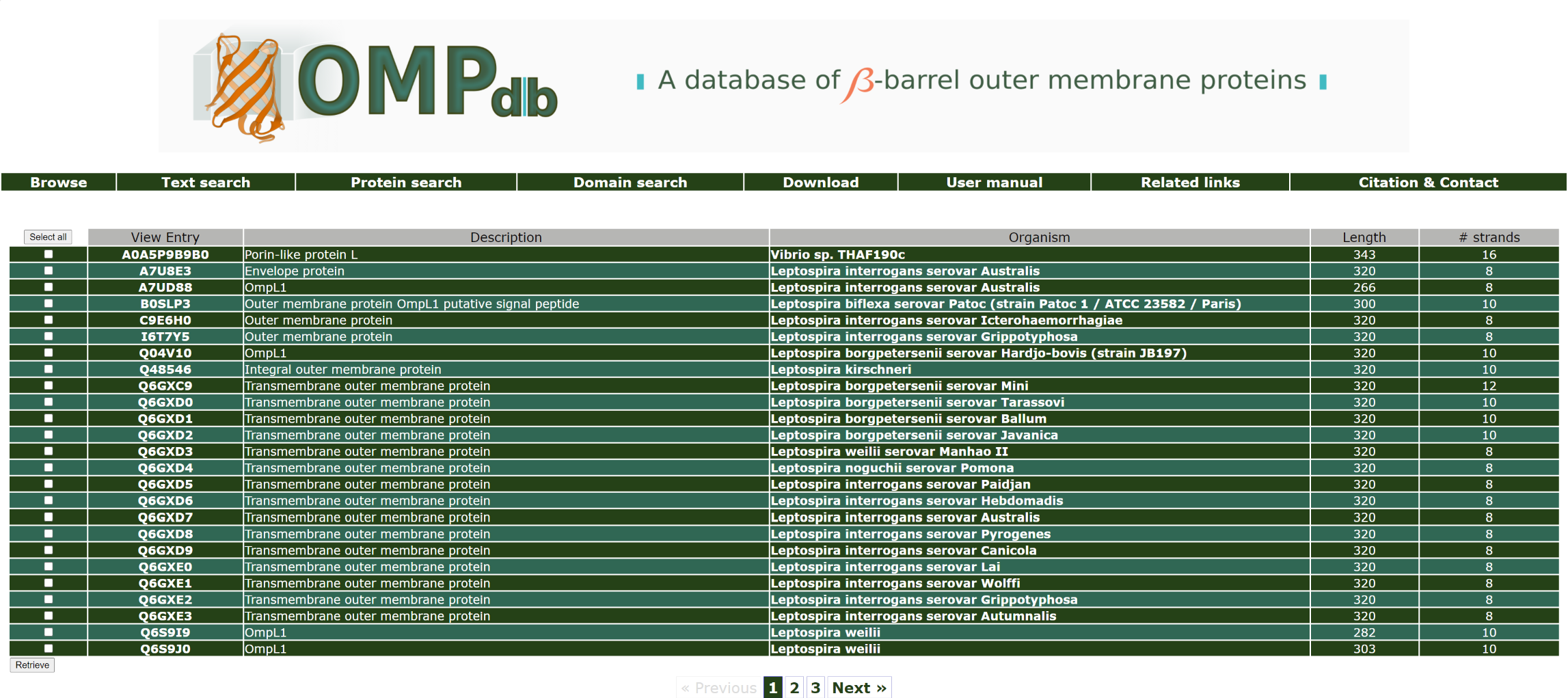
Return to top
Organism name
Corresponds to the field Species of an entry.
Example 3:
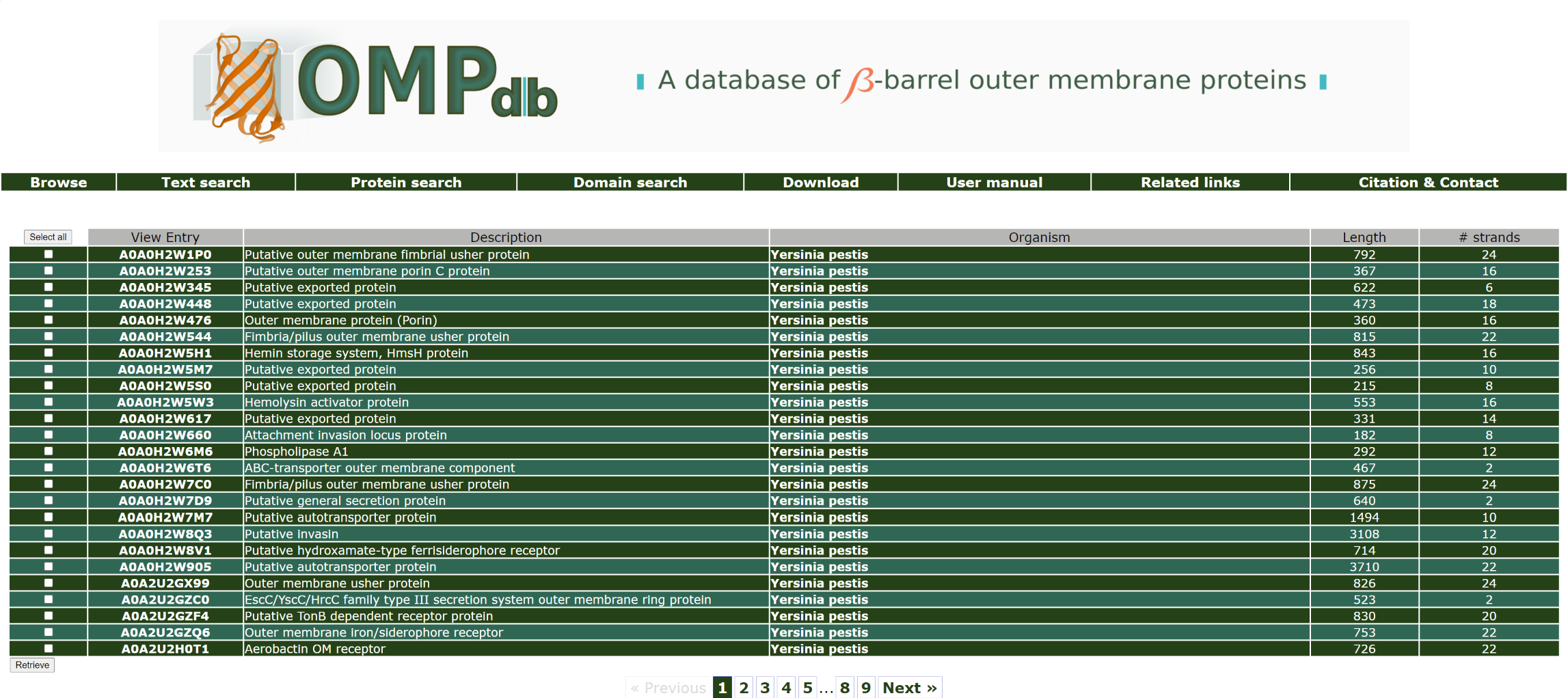
If the user wants to retrieve all proteins that originate from species Geobacter, he/she has to enter Geobacter as a query in the Organism name field:
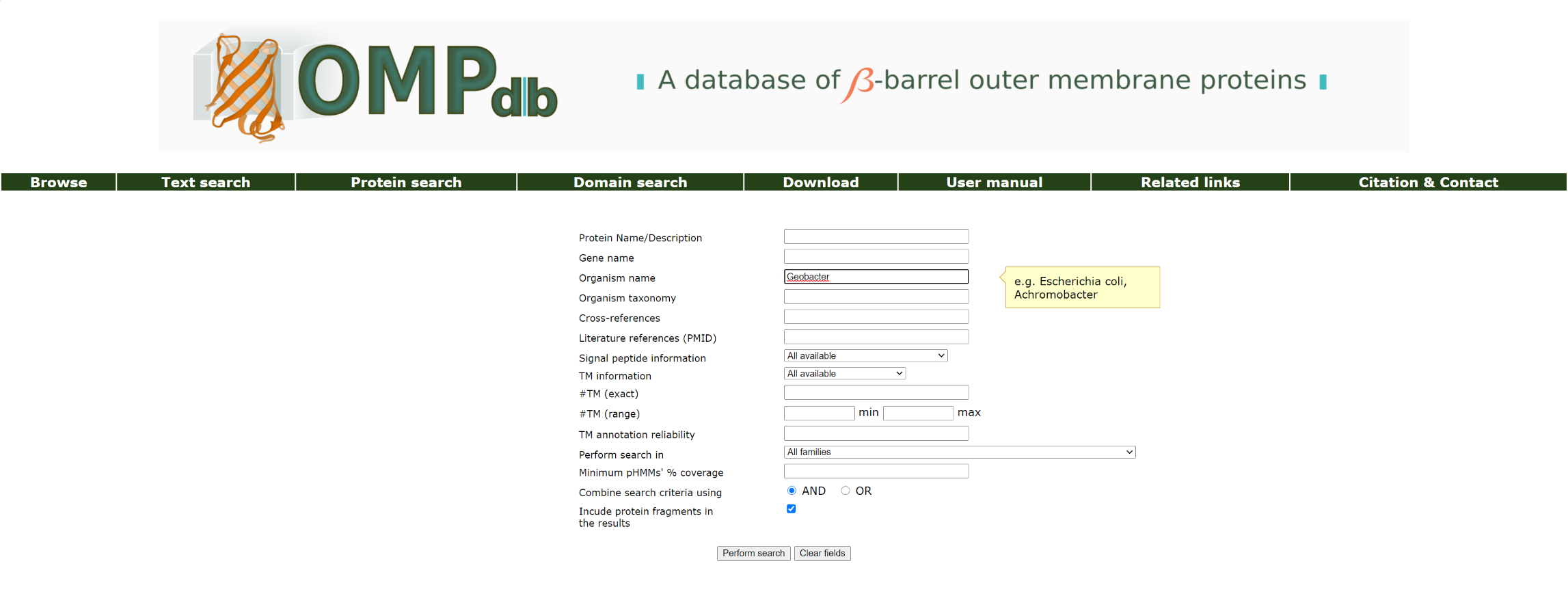
The top of the result page for the above search is:
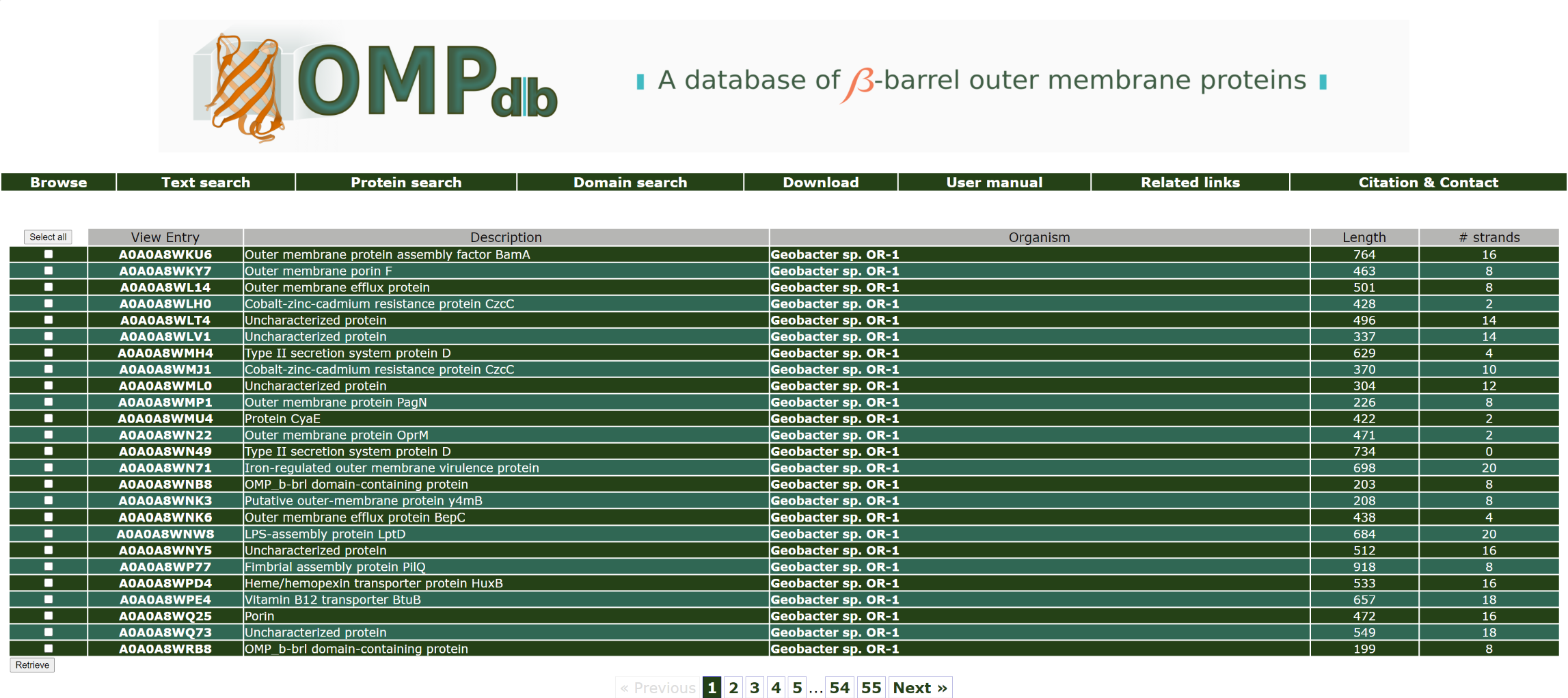
Return to top
Organism taxonomy
Corresponds to the field NCBI taxonomy of an entry.
Example 4:
If the user knows the NCBI taxonomy number, he/she can search with it as a keyword in the Organism taxonomy field:
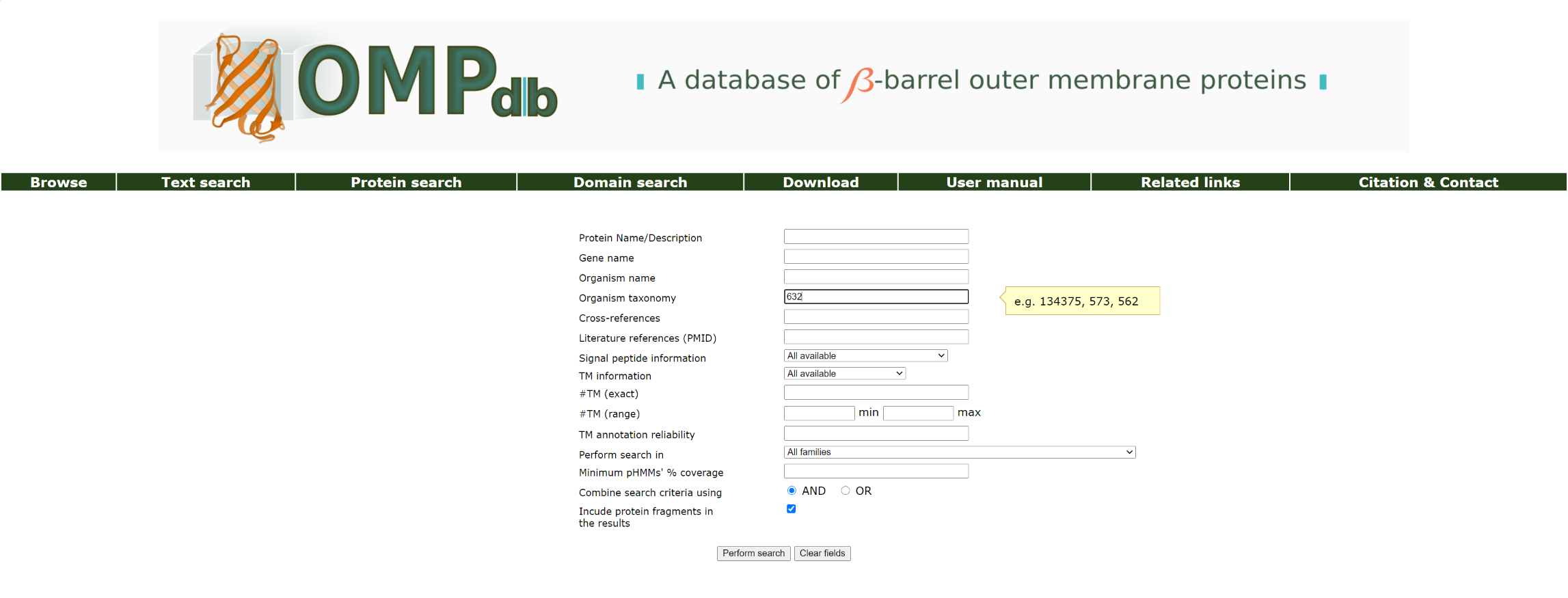
The top of the result page for the above search is:
Return to top
Cross-references
Corresponds to the field Cross-references of an entry and in specific to the accession numbers or IDs from other selected publicly accessible biological databases worldwide.
Example 5:
If the user wants to retrieve a protein included in OMPdb and has a given Uniprot accession number or ID of any other database that is cross-linked in OMPdb (namely PFAM, InterPro, ProDom, PRINTS, PDB, EMBL, SMART, PROSITE, PIR, Psortdb), he/she has to enter it as a query in the Cross-references field.
For example in order to retrieve proteins included in OMPdb that have the InterPro code IPR023614, the user has to use the name IPR023614 as a query:
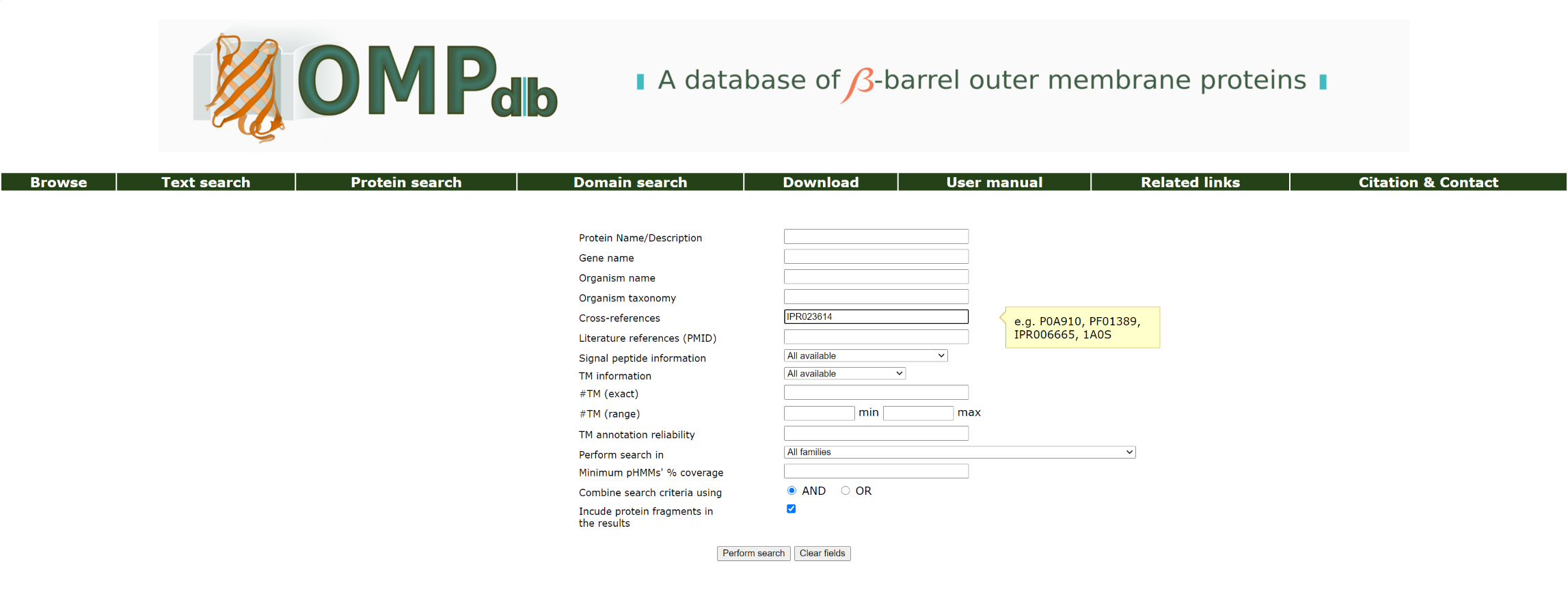
The top of the result page for the above search is:
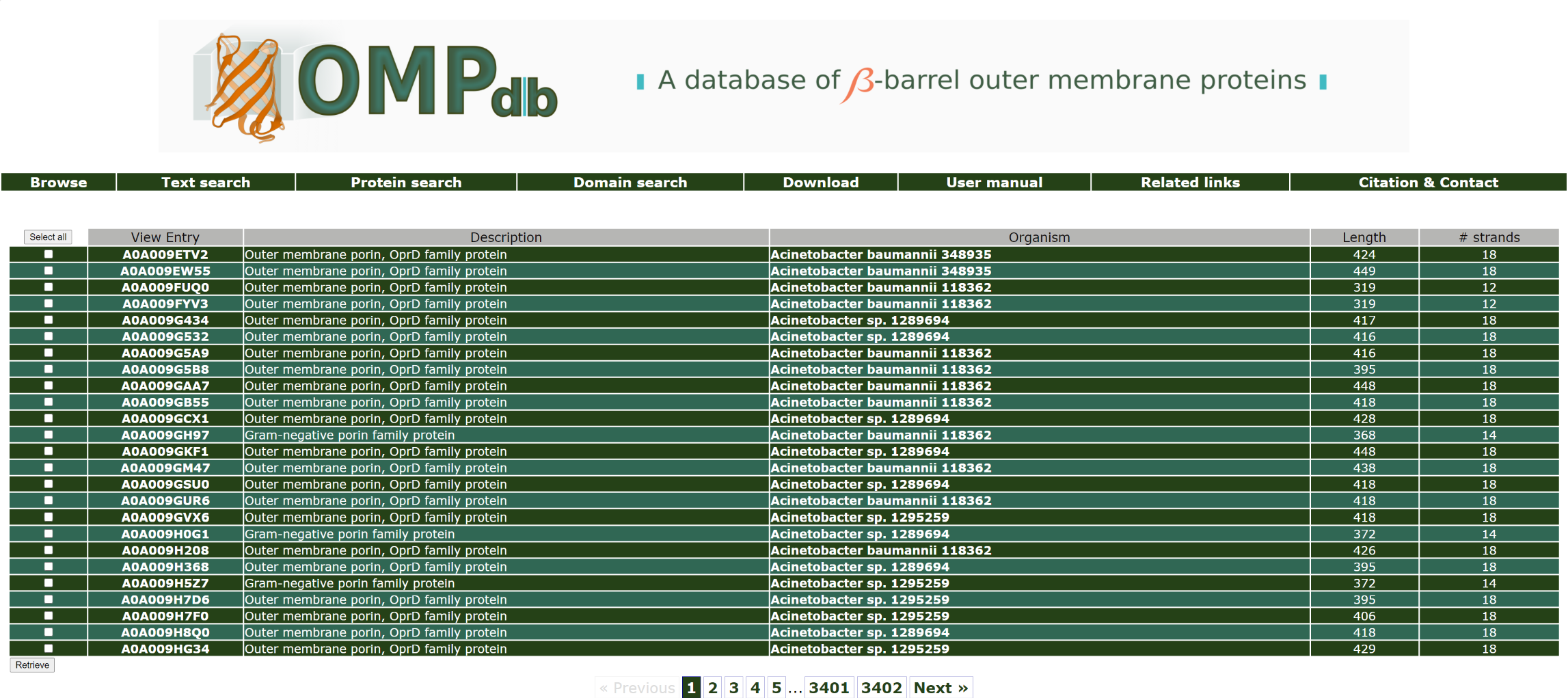
Return to top
Literature references (PMID)
Here the user can retrieve entries from the database that are associated with a particular literature reference by entering the respective Pubmed ID.
Example 4:
We perform a text search using the Pubmed id 1406271 in the Literature references (PMID) field:
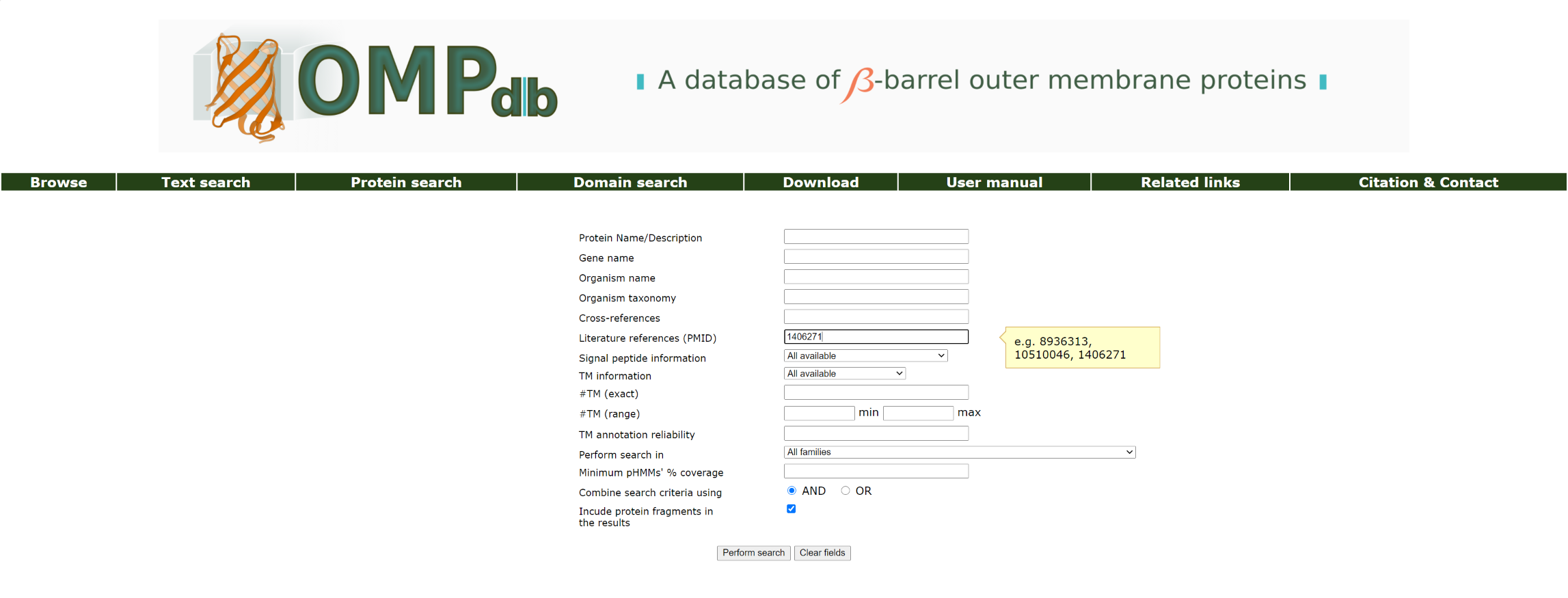
The top of the result page for the above search is:
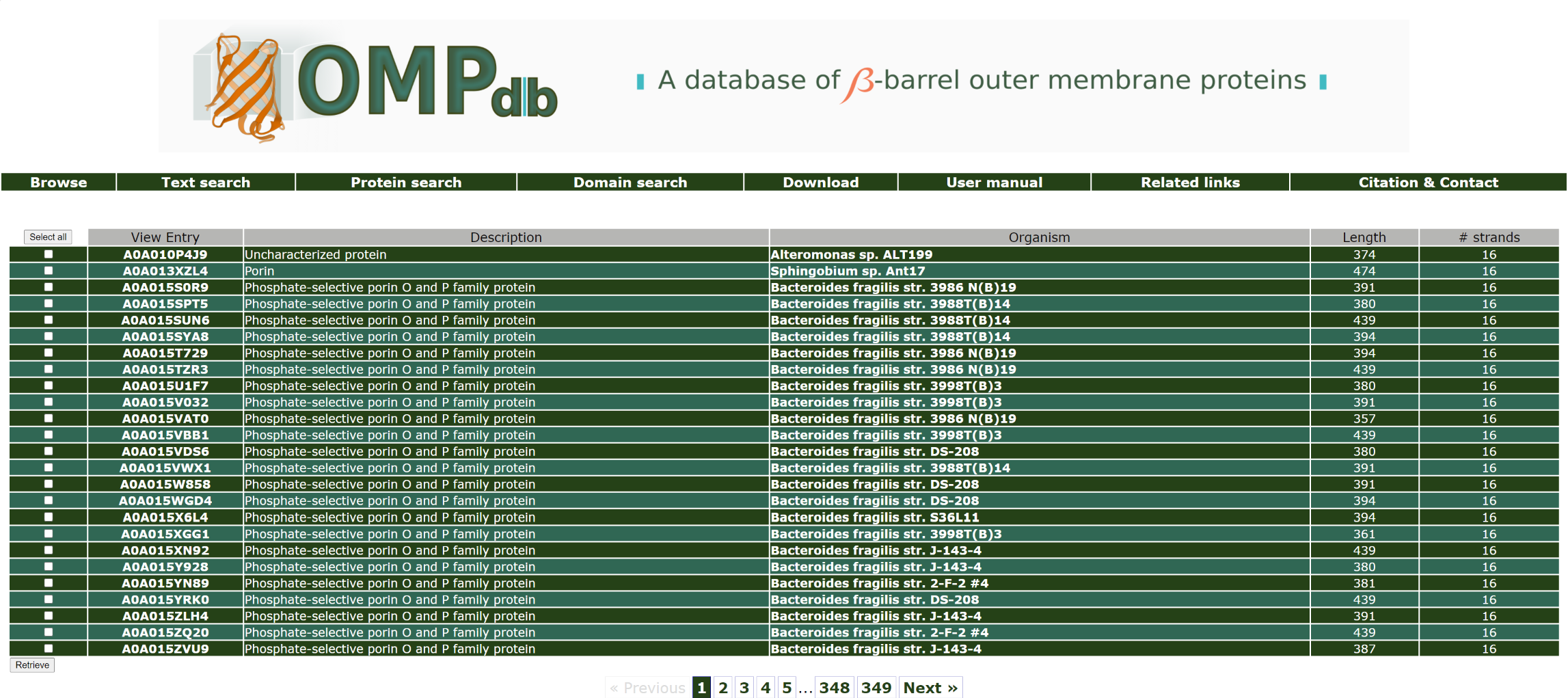
Return to top
Combining search criteria
The user can combine each of the above-mentioned criteria with the information provided in the two drop-down menus of the webpage. Information concerning the presence of the signal peptide can be set, either originating from Uniprot (Experimentally verified or Experimentally verified [by similarity]) or based on the results from SignalP ver. 5.0 predictions.
Example 6:
If the user wants to retrieve all proteins with protein name containing porin, that have Experimentally verified signal peptide, their beta strands annotation is homologous to a 3D-structure, they contain between 8 and 20 beta strands and originate from Geobacter, while covering at least 82% of the family's profile HMM, he/she must fill in the appropriate fields and click AND in order to perform this combined search:
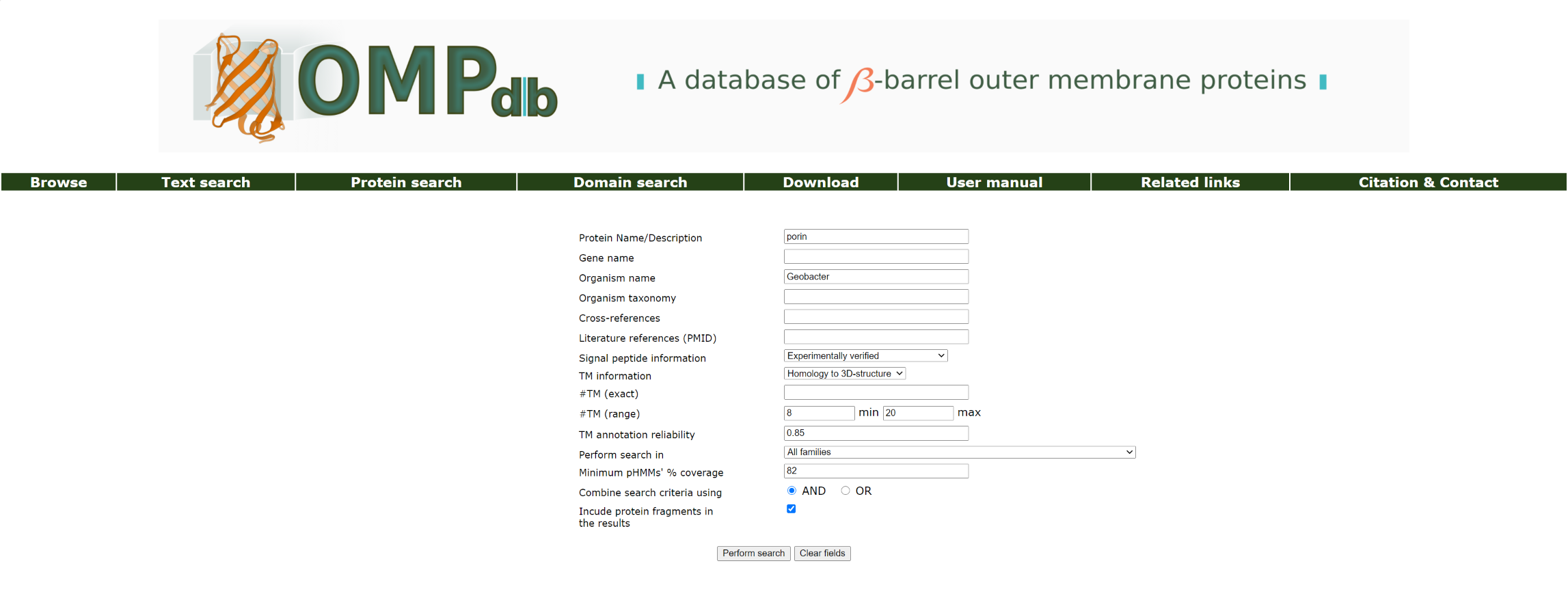
The top of the result page for the above search is:
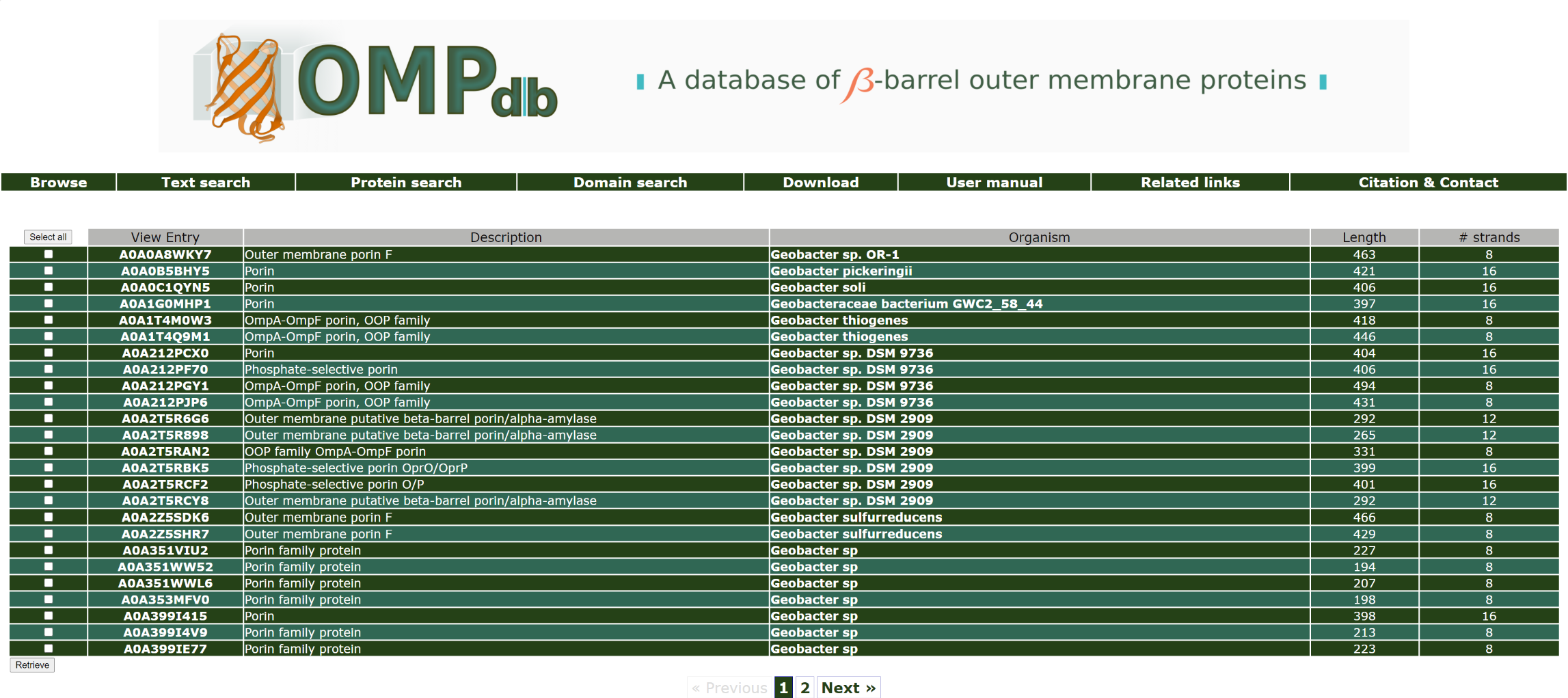
Select and download entries
In every case, the user can retrieve one or more or the protein entries that are presented in the
Text search result page in various formats (flat text,
fasta files or XML). This can be done by clicking on the checkbox that is on the left of each protein result and then pressing
the Retrieve button as shown in the picture below:
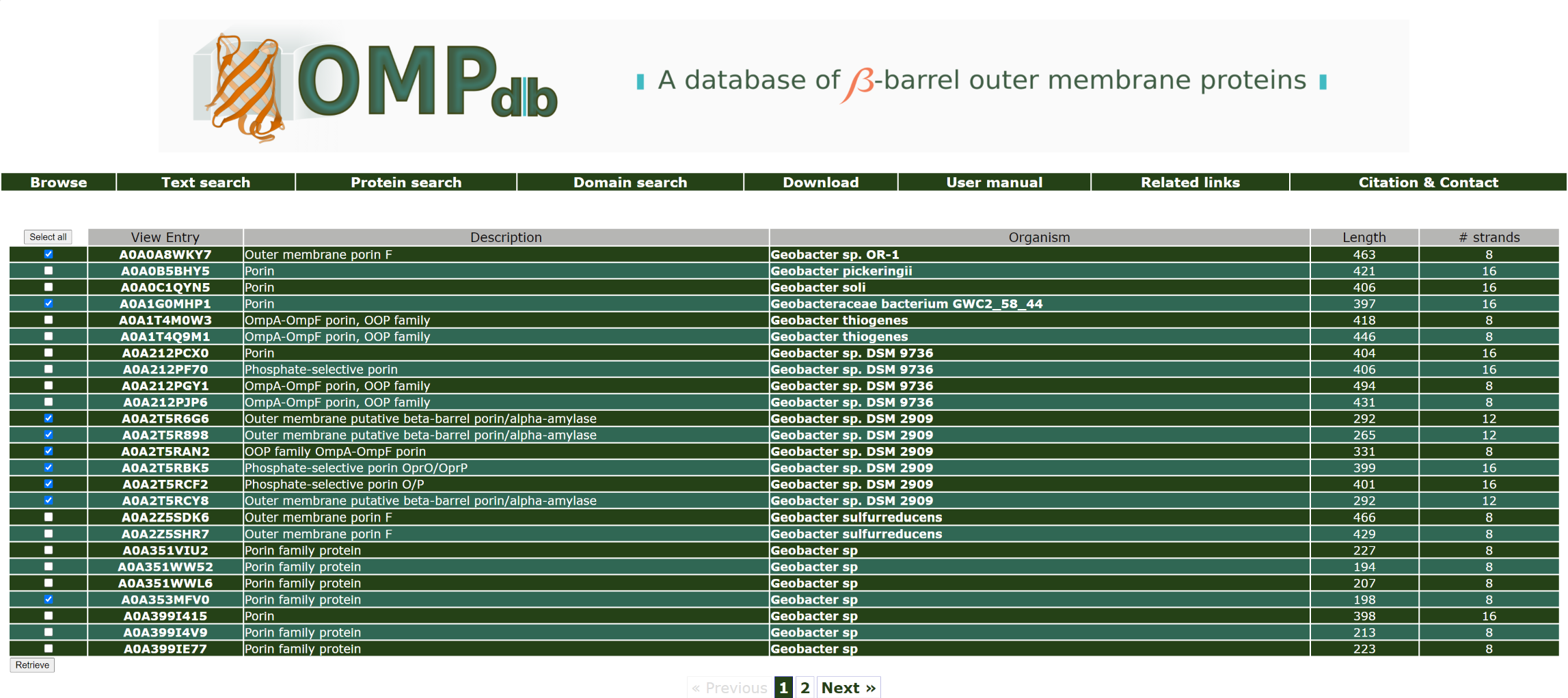
By doing so, the user will be redirected to the page from which he/she can download the selected protein entries in one of the
provided formats:
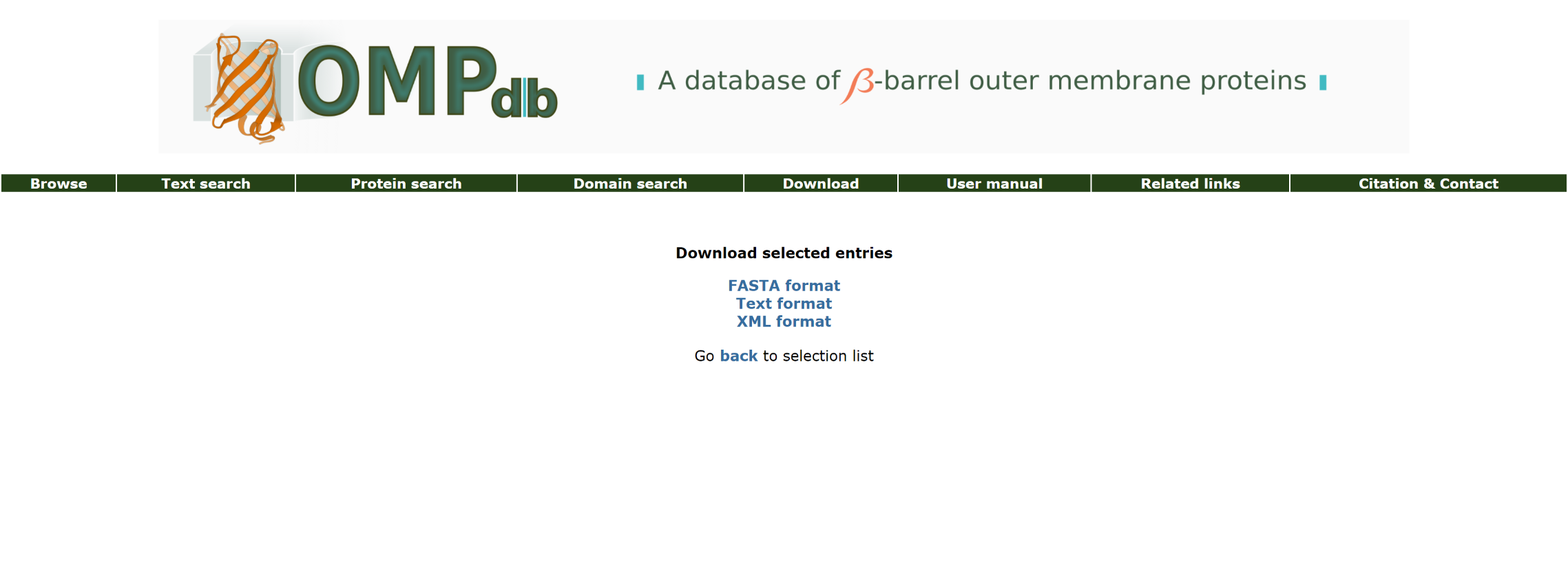
Return to top
Protein search
With the Protein search tool, the user may submit a sequence and search the database for sequence homologues. The input for the BLAST search application is a FASTA-formated protein sequence (either pasted in the textarea provided or uploaded as a text file). The user can select between performing a typical BLAST search or a phmmer search:
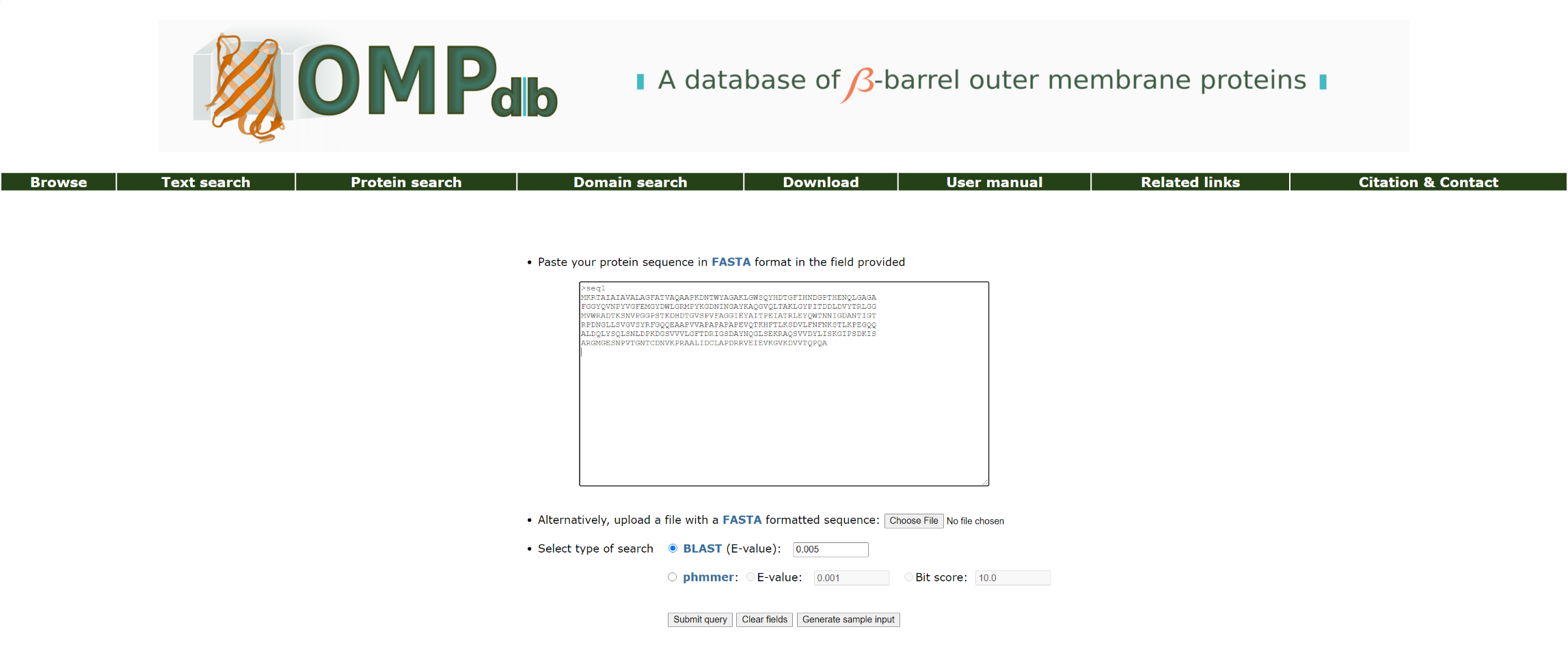
Figure 4. Basic view of the Protein search page.
The result page of the Protein search when BLAST is selected shows a list of the significant hits, along with the query sequence the user has submited. The list is in a table format including the Uniprot Accession number of the target protein, Length of the target sequence and the Query and Target align range. The BLAST results can be compared through the Score and E-value and the Identities, Positives and Gaps as well. The input and output files are also at the user's disposal.
The top of the result page for the above search is:
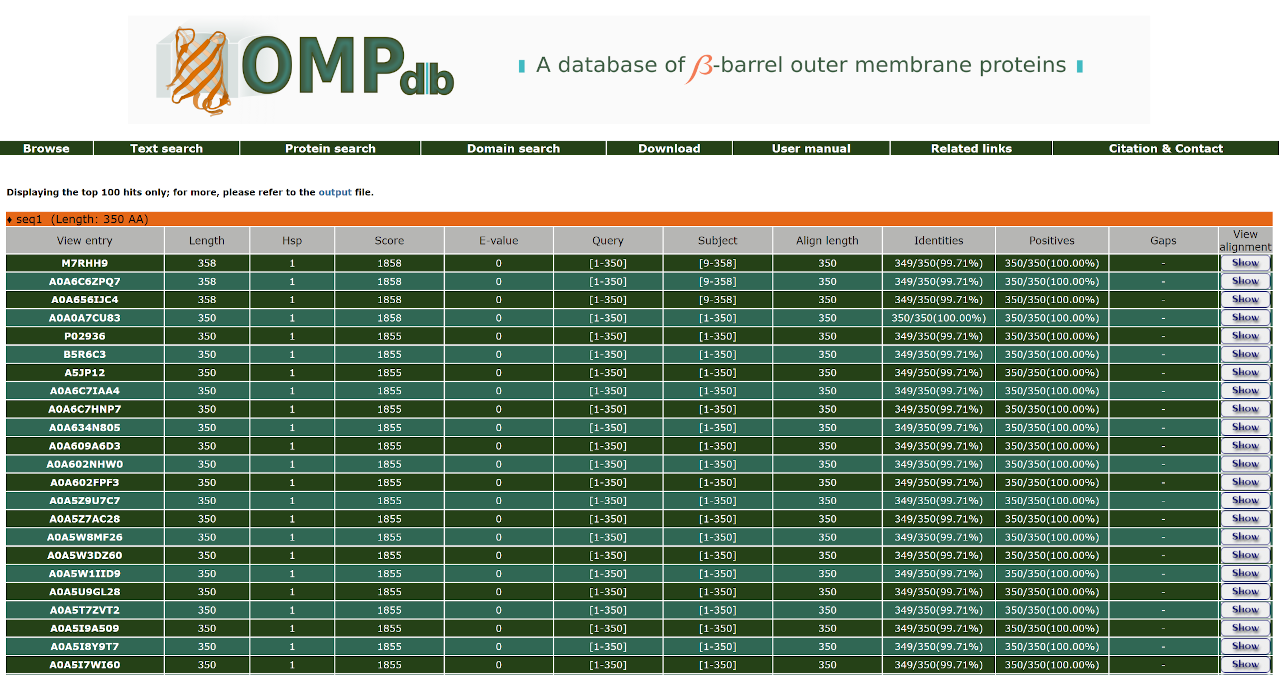
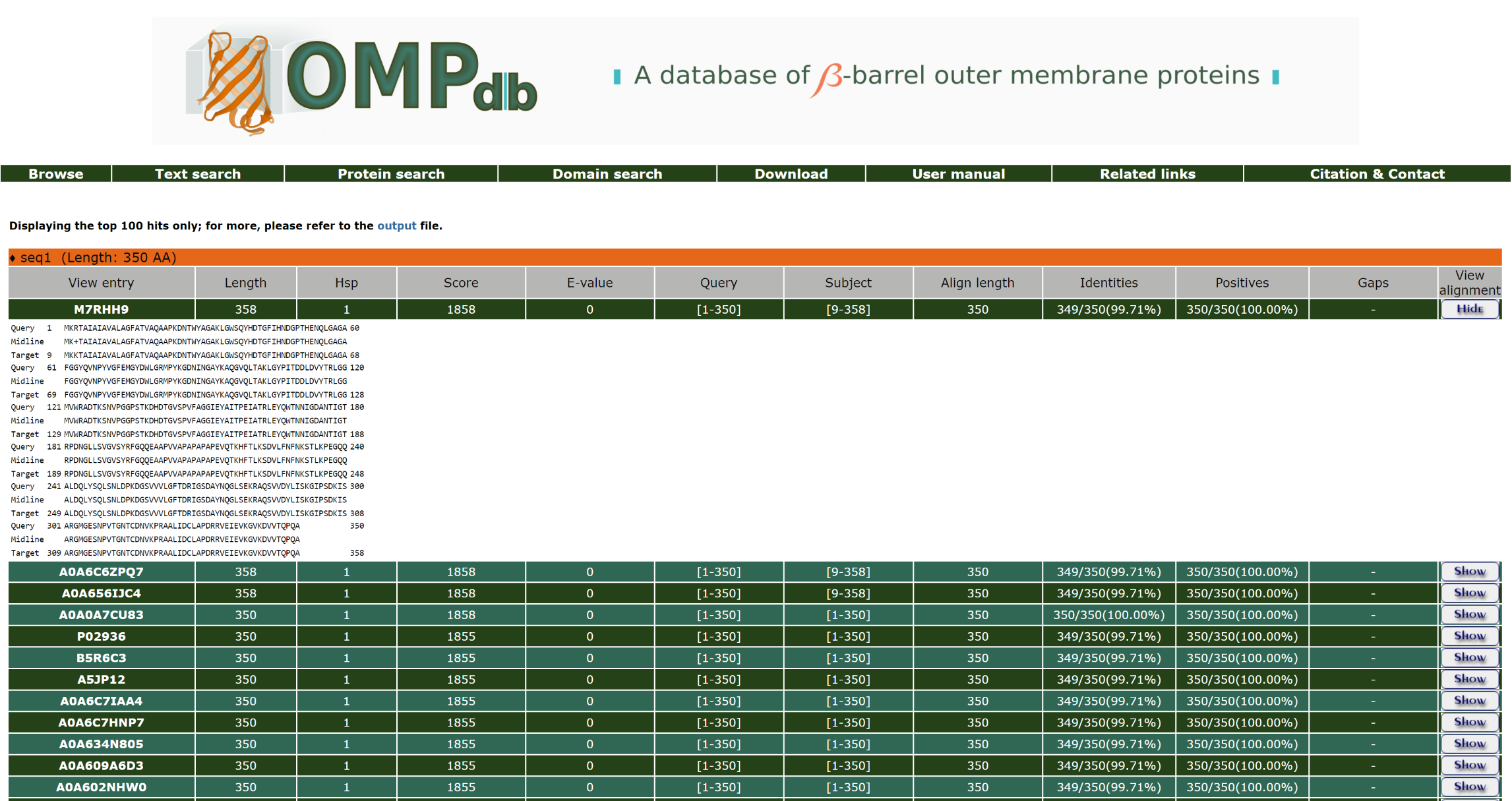
Furthermore, the user can have a more detailed view of each alignment through the Show/Hide button at the end of each line:
The result page of the Protein search when pHMMER is selected shows a list of the significant hits, along with the query sequence the user has submited. The list is in a table format including the Uniprot Accession number of the target protein and the sequence coverage lengths of both the Query and Target sequences. The pHMMER results can be compared through the Score and E-value. The input and output files are also at the user's disposal.
The top of the result page for the above search is:
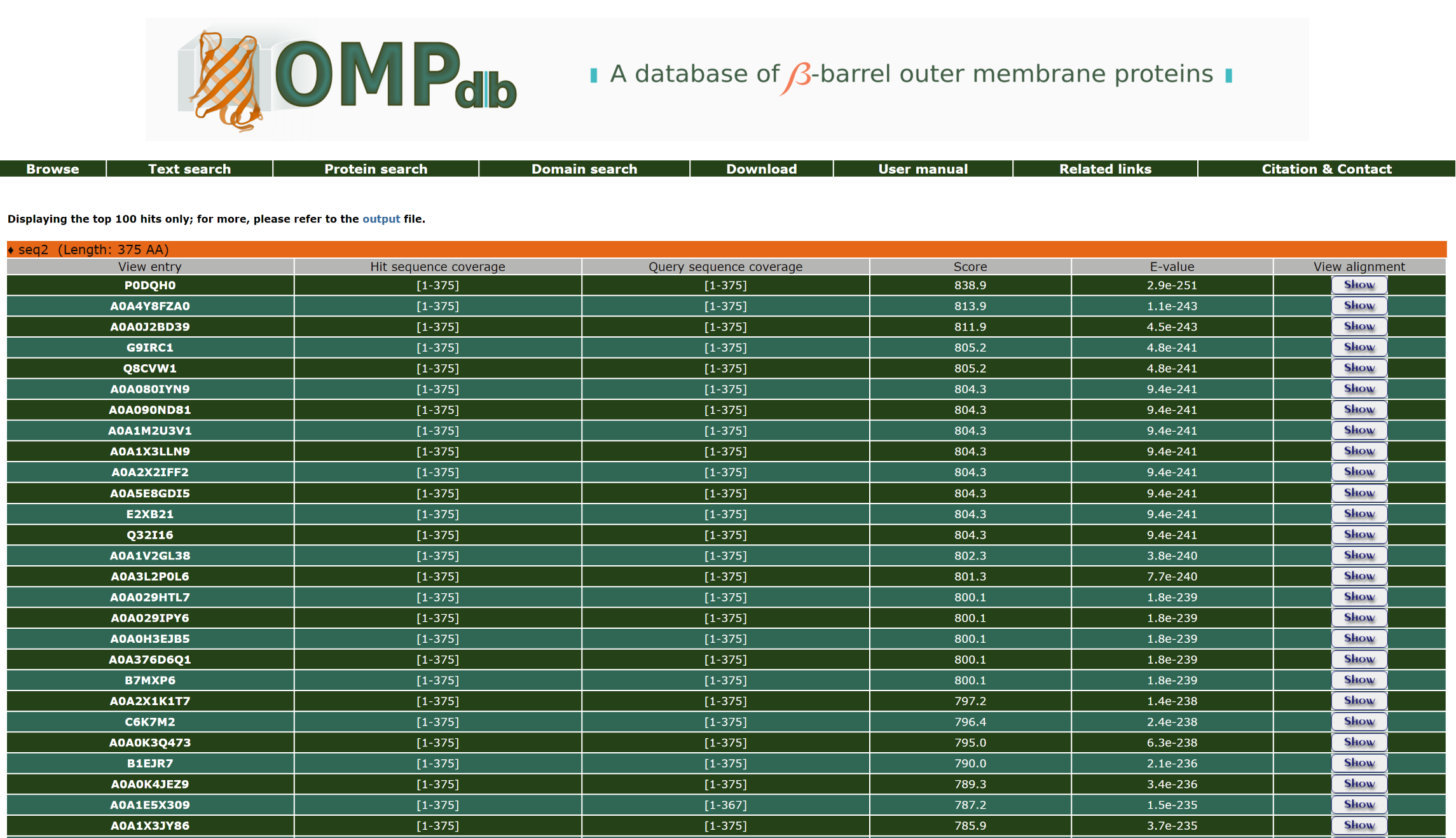
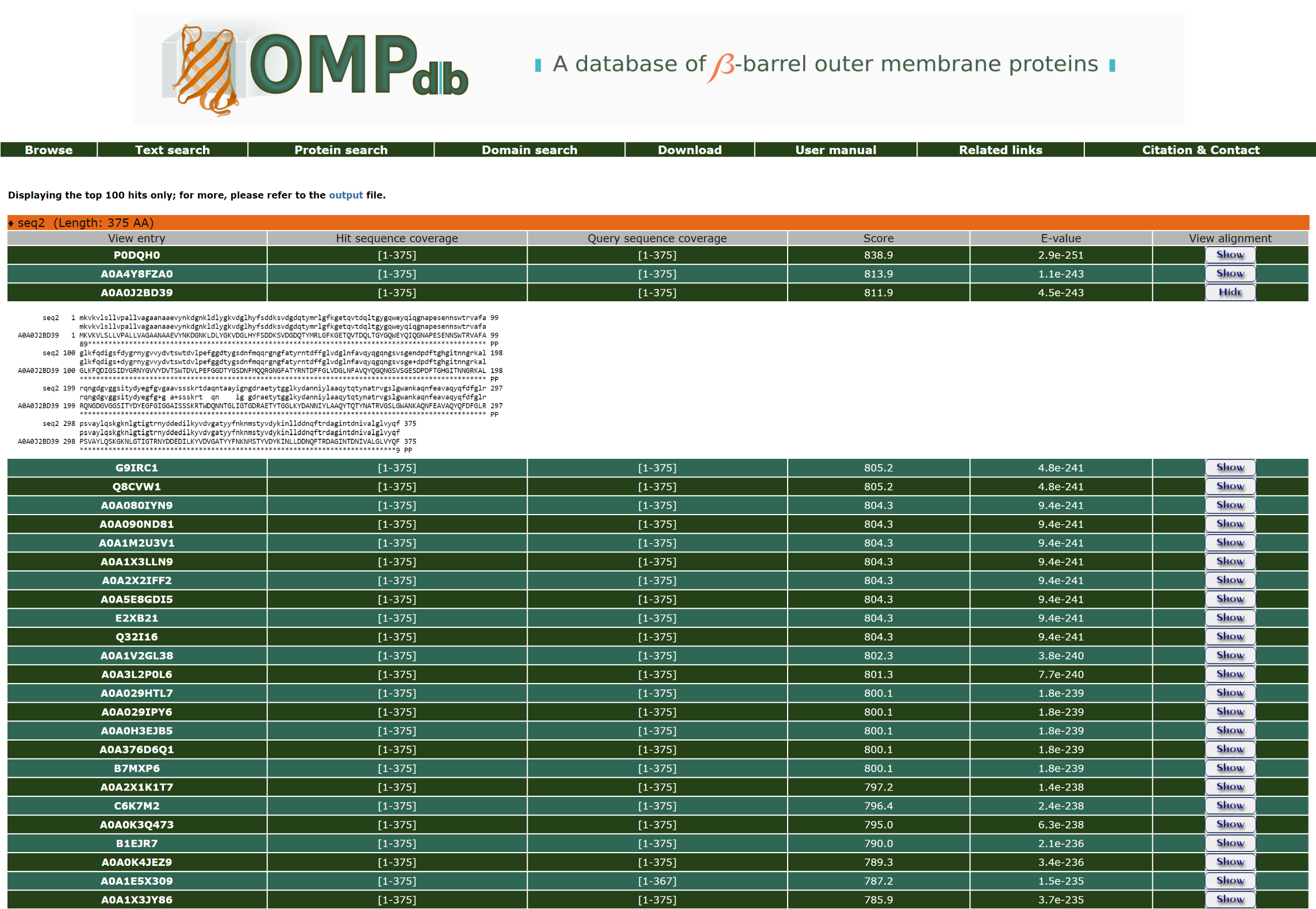
Furthermore, the user can have a more detailed view of each alignment through the Show/Hide button at the end of each line:
Return to top
Domain search
With the Domain search tool, the user may submit one or more sequences (up to 200 each time) to be searched against the OMPdb's collection of pHMMs. The input for the domain search application is again FASTA-formated sequences. If there is a significant hit in one of OMPdb's pHMMs, the input sequence(s) will be classified in one of the available families. The user can specify an E-value or Score cutoff level if he/she wishes or rely on the Gathering threshold or Trusted cutoff which characterize each model.
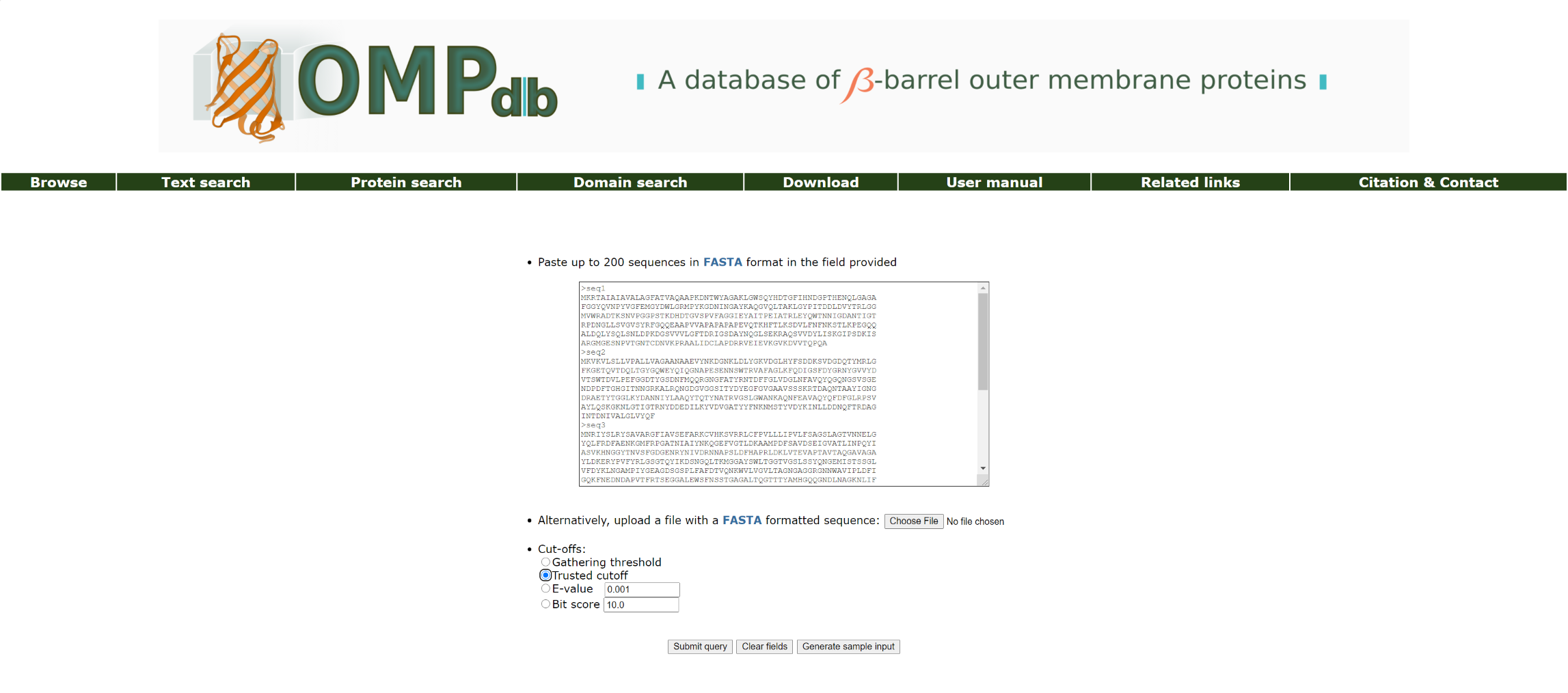
Figure 5. Basic view of the domain search page.
The result page of the domain search shows a tabular output which includes the Respective OMPdb family of the hit (if any), the Sequence and Model coverage and, finally, the Score and E-value of the hits as well. The results are listed from the lower to the highest E-value so that the significant hits are separated from the insignificant ones. The input and output files are also at the user's disposal for further analysis.
A part of the result page for the above domain search is:
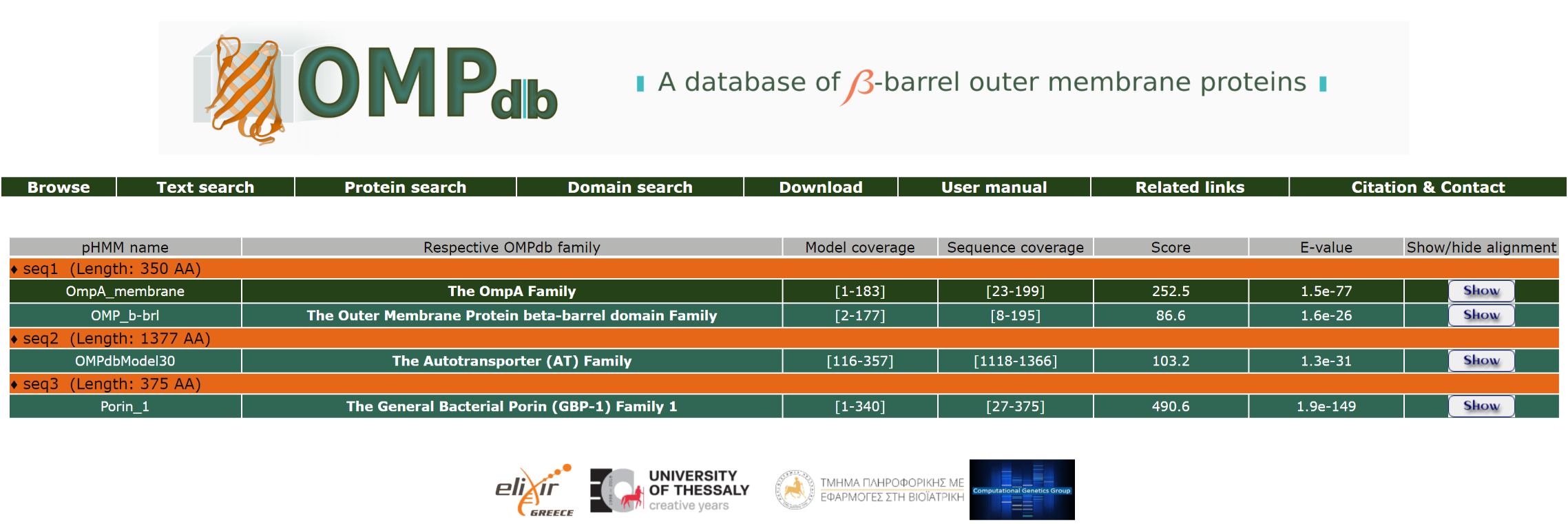
Furthermore, the user can have a more detailed view of the alignment through the Show/Hide button at the end of each line:
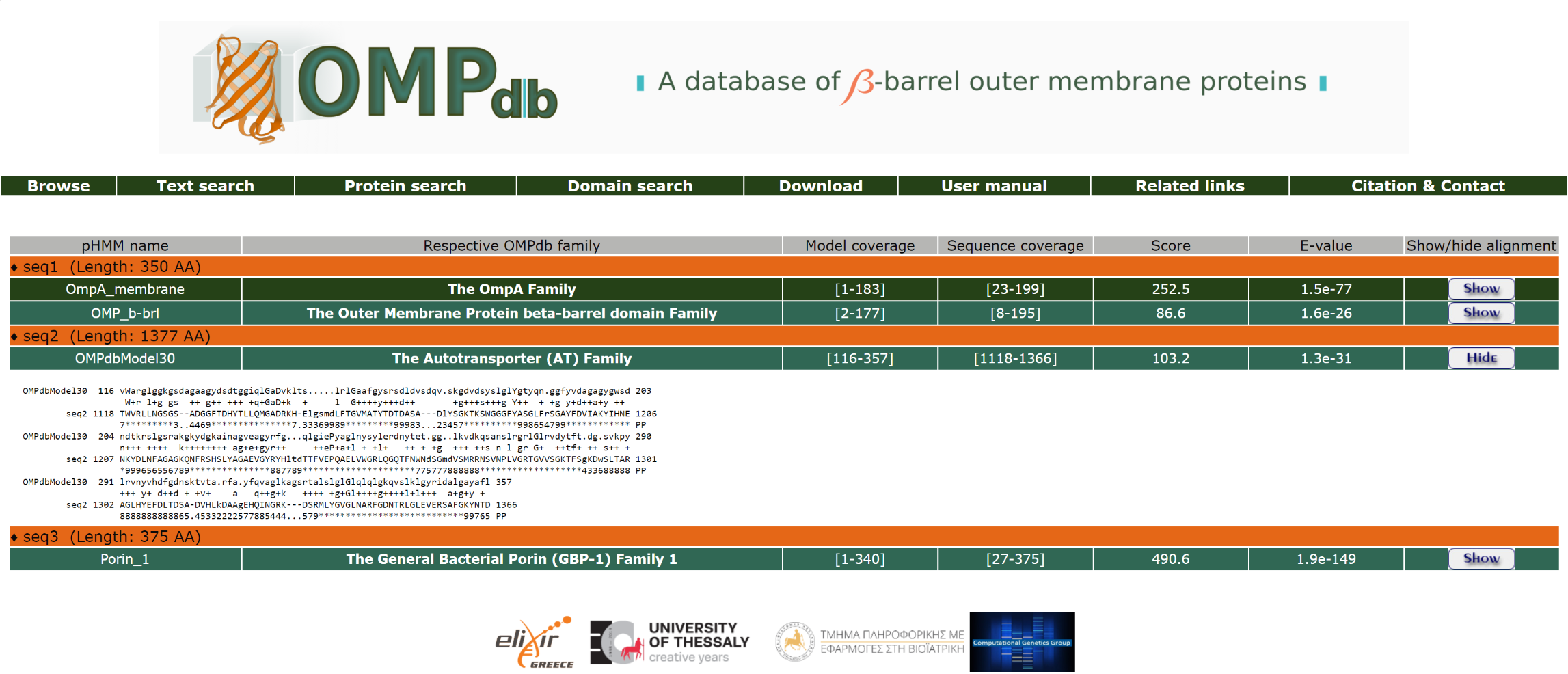
Return to top
Detailed view of an entry
Each entry in OMPdb has an unique OMPdbID number which is presented at the top of each entry (e.g. ::OMPdbID1982::). Furthermore, the user can choose from other views of the entry besides of the basic view (FASTA, Raw text and XML). More details about these formats are described in the Download section of this manual. If there is an available file with PDB coordinates for the structure of the protein (based on homology modeling), it is also available for download.
The available fields in a protein entry of OMPdb are organized into four sections:
First, there is the Protein details section which contains fields describing general information about the protein and are collected mainly from Uniprot. These include:
- OMPdb Family
- Protein description
- Gene name
- Species
- NCBI taxonomy
- Protein sequence
- Sequence length
- Fragment: this field informs whether the protein is a fragment of not.
- pHHM coverage: shows the coverage of the profile HMM model in regards to the protein.
By clicking on the organism's name, the user can get a list of all proteins in OMPdb that belong to this organism.
The Cross references section which follows contains fields corresponding to accesion numbers that link to other public available databases which contain information for the protein such as:
- Uniprot
- PFAM
- InterPro
- ProDom
- PRINTS
- PDB
- EMBL
- SMART
- PROSITE
- PIR
The Signal Peptide Information section contains fields that provide information concerning the existence of a signal peptide:
- Amino acids: the start and end amino acids of the signal peptide in the protein sequence.
- Sequence: the sequence of the signal peptide alone.
-
Annotation source: the source of information about the existence of the signal peptide [experimentally verified, experimentally verified by similarity or prediction methods (SignalP 5.0].
The TM annotation section refers to the existing information on the transmembrane (TM) segments (beta strands) of the protein (if any):
There are 3 possibilities:
- if there is a 3D structure available for the given protein, the annotation of the TM segments is based on the information from the PDBTM database.
- if the protein belongs to the same family which includes a protein with experimentally verified annotation of TM segments, then this information is mapped on the sequence of this protein as well using the full alignment of the families' sequences.
- if the number of TM segments that are deduced using the strategy in step #2 is not equal to the number of TM segments that is known for the members of the corresponding family, or if there is no annotation for the TM segments whatsoever, then the the TM-annotation is based on the PRED-TMBB2 algorithm.
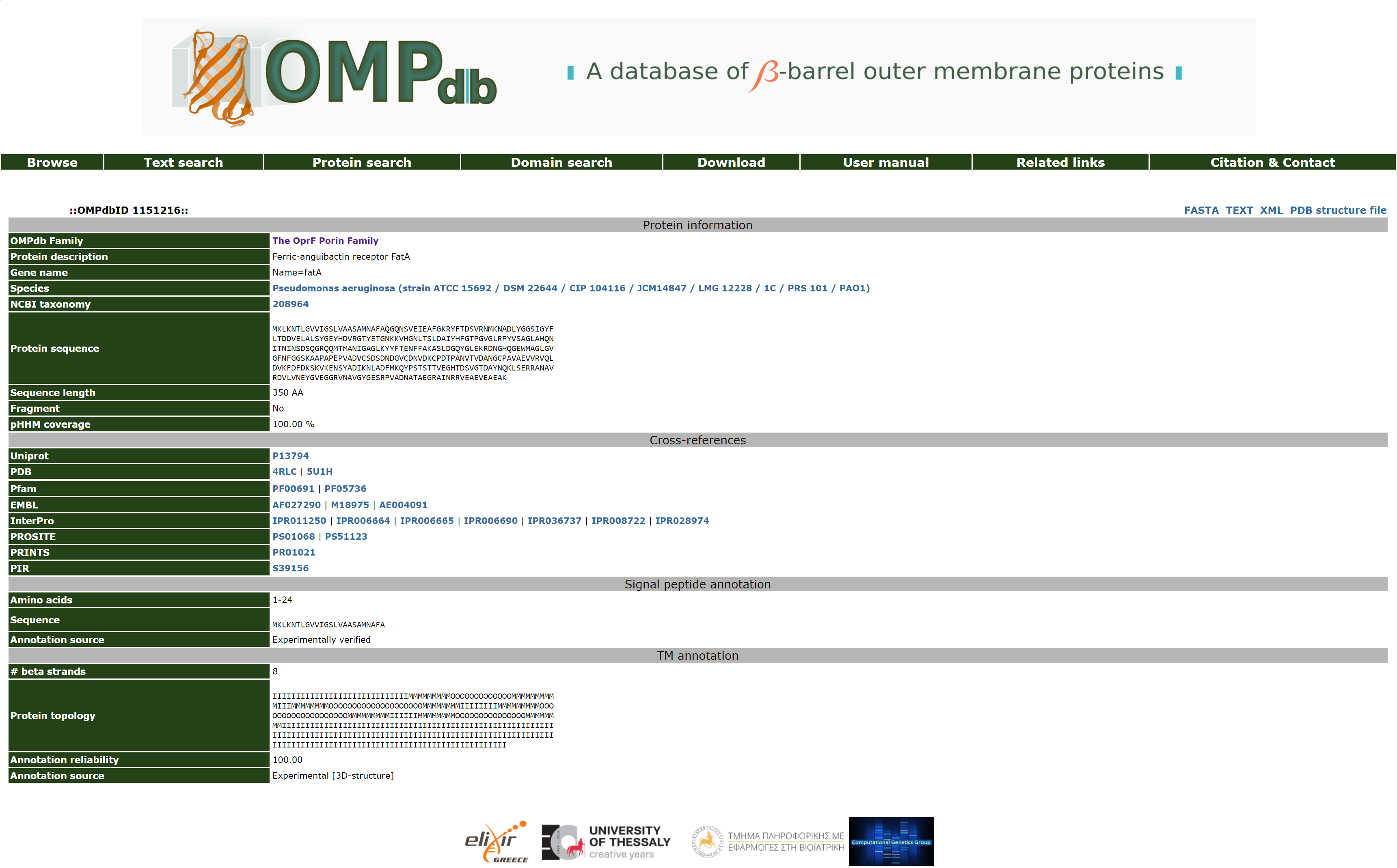
Figure 6. Detailed view of an OMPdb entry.
Return to top
Download
Through the Download page, the user can download the most up-to-date version of the database in one of the following formats:
Our target is to update OMPdb frequently, in order to keep it synchronized with the regular updates of the Uniprot database (usually once per month) and the deposited 3D-structures of ß-barrel outer membrane proteins as well.
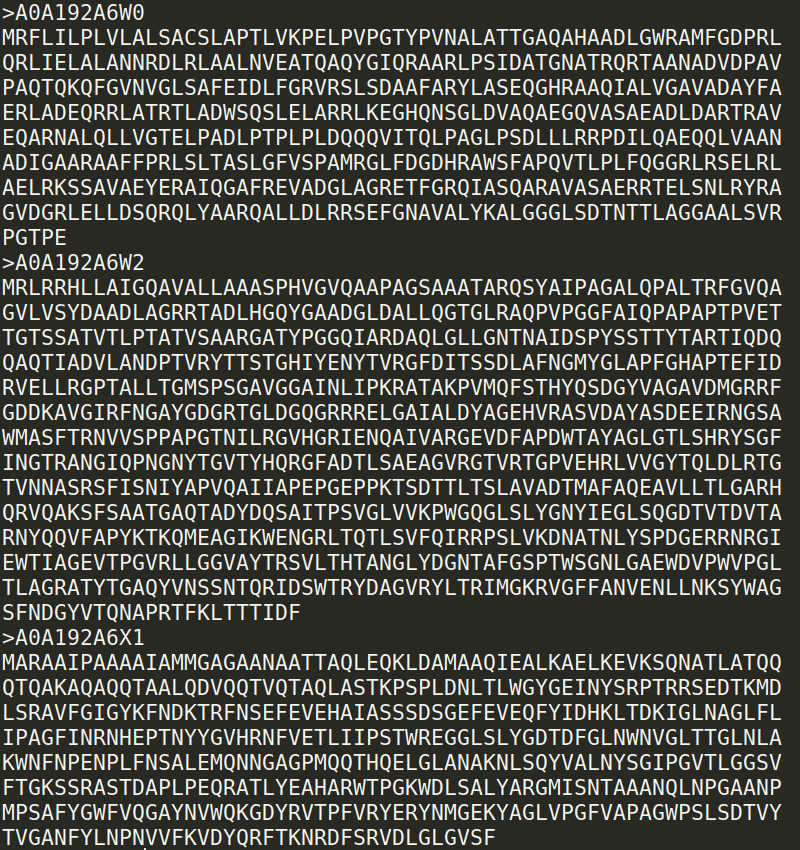
The OMPdb FASTA format corresponds to the sequences of all proteins in OMPdb in FASTA format.
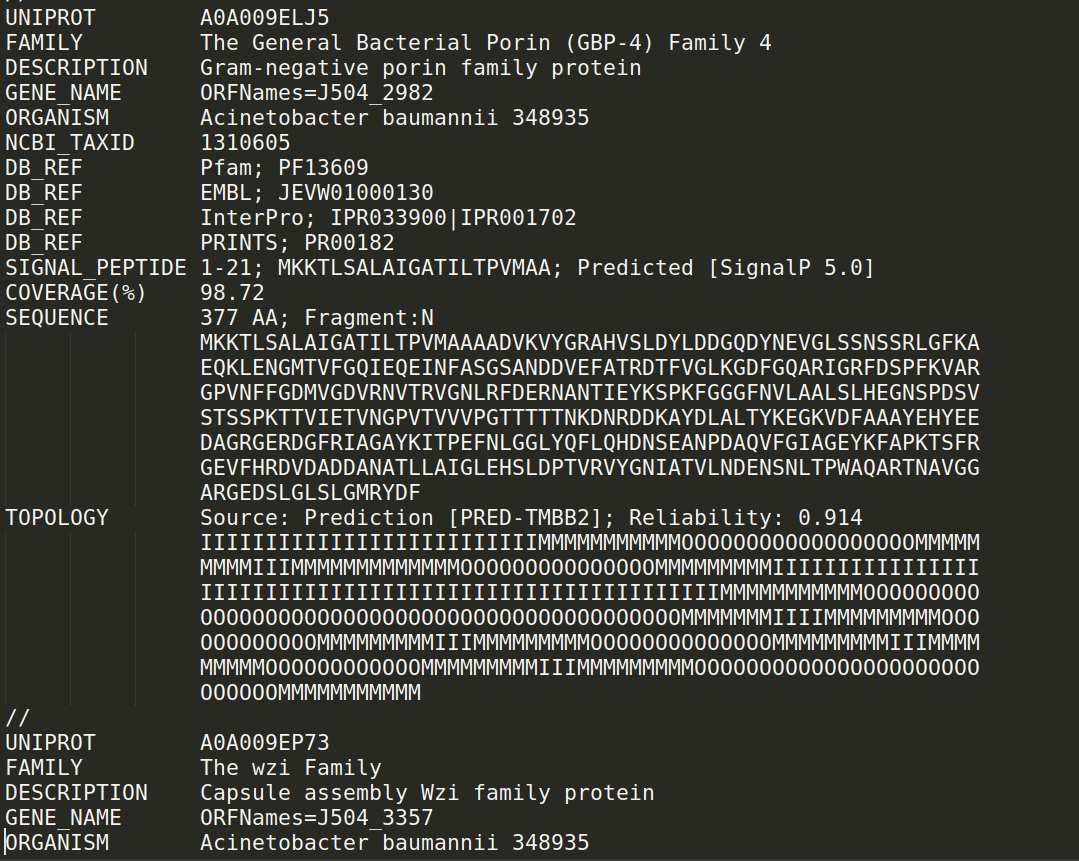
The OMPdb Text format corresponds to all entries in OMPdb in plain text format. Entries are separated by "//" and all information contained in a typical OMPdb entry is encoded in raw text format allowing easier and faster data manipulation:
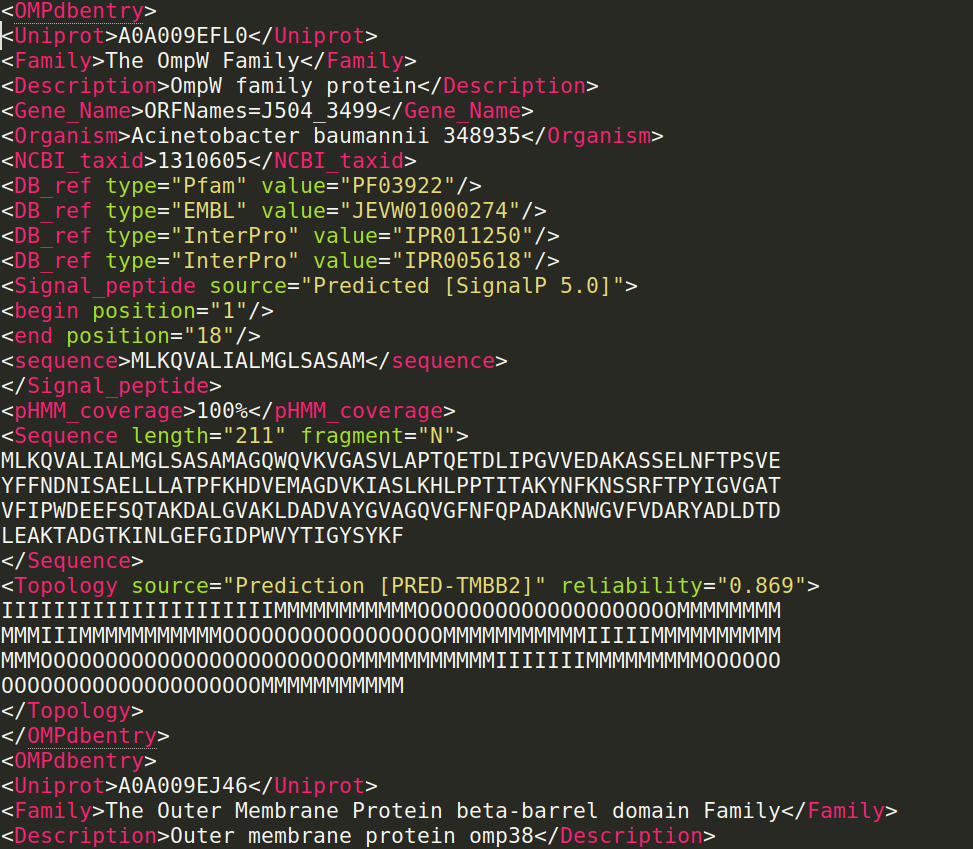
The OMPdb XML format corresponds to all entries in OMPdb in Extensible Markup Language (XML) format:
Return to top